Circular RNA
Circular RNA (or circRNA) is a type of single-stranded RNA which, unlike linear RNA, forms a covalently closed continuous loop. In circular RNA, the 3' and 5' ends normally present in an RNA molecule have been joined together. This feature confers numerous properties to circular RNA, many of which have only recently been identified.
Many types of circular RNA arise from otherwise protein-coding genes. Some circular RNA has been shown to code for proteins.[1][2] Some types of circular RNA have also recently shown potential as gene regulators. The biological function of most circular RNA is unclear.
Because circular RNA does not have 5' or 3' ends, it is resistant to exonuclease-mediated degradation and is presumably more stable than most linear RNA in cells.[3] Circular RNA has been linked to some diseases such as cancer.[4]
RNA splicing
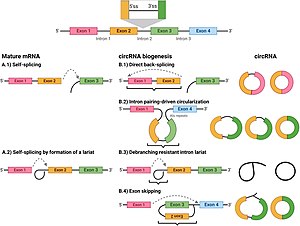
In contrast to genes in bacteria, eukaryotic genes are split by non-coding sequences called introns. In eukaryotes, as a gene is transcribed from DNA into a messenger RNA (mRNA) transcript, intervening introns are removed, leaving only exons in the mature mRNA, which can subsequently be translated to produce the protein product.[5] The spliceosome,[5] a protein-RNA complex located in the nucleus, catalyzes splicing in the following manner:
- The spliceosome recognizes an intron, which is flanked by specific sequences at its 5' and 3' ends, known as a donor splice site (or 5' splice site) and an acceptor splice site (or 3' splice site), respectively.
- The 5' splice site sequence is then subjected to a nucleophilic attack by a downstream sequence called the branch point, resulting in a circular structure called a lariat.
- The free 5' exon then attacks the 3' splice site, joining the two exons and releasing a structure known as an intron lariat. The intron lariat is subsequently de-branched and quickly degraded.[5]
Alternative splicing
Alternative splicing is a phenomenon through which one RNA transcript can yield different protein products based on which segments are considered "introns" and "exons" during a splicing event.[5] Although not specific to humans, it is a partial explanation for the fact that humans and other much simpler species (such as nematodes) have similar numbers of genes (in the range of 20 - 25 thousand).[6] One of the most striking examples of alternative splicing is in the Drosophila DSCAM gene, which can give rise to approximately 30 thousand distinct alternatively spliced isoforms.[7]
Non-canonical splicing
Exon scrambling
Exon scrambling, also called exon shuffling, describes an event in which exons are spliced in a "non-canonical" (atypical) order. There are three ways in which exon scrambling can occur:
- Tandem exon duplication in the genome, which often occurs in cancers.
- Trans-splicing, in which two RNA transcripts fuse, resulting in a linear transcript containing exons that, for example, may be derived from genes encoded on two different chromosomes. Trans-splicing is very common in C. elegans
- A splice donor site being joined to a splice acceptor site further upstream in the primary transcript, yielding a circular transcript.[8]
The notion that circularized transcripts are byproducts from imperfect splicing is supported by the low abundance and the lack of sequence conservation of most circRNAs,[9] but has been challenged.[8][10][3]
Alu elements impact circRNA splicing
Repetitive Alu sequences represent approximately 10% of the human genome.[11] The presence of Alu elements in flanking introns of protein-coding genes adjacent to the first and last exons that form circRNAs, influence the formation of circRNAs.[12][13][3][14] It is important that the flanking intronic Alu elements are complementary, as this enables RNA pairing, which in turn facilitates circRNA synthesis.[15]
Impact of RNA editing on circRNA formation
RNAs can undergo base modification by RNA editing after transcription. RNA editing occurs mainly in Alu elements of protein-coding genes.[16] A-to-I RNA editing in up- and downstream intronic Alu elements flanking the back-splice site (BSS) can reduces the formation of circRNAs in the human heart.[16] In the failing human heart, a predominant reduction in A-to-I RNA editing leads to an increased formation of circRNAs, which is presumably mediated by better complementary pairing of RNA of the Alu elements flanking the back-splice site.[16]
Characteristics of circular RNA
Early discoveries of circRNAs
Early discoveries of circular RNAs led to the belief that they lacked significance due to their rarity. These early discoveries included the analysis of genes like the DCC and Sry genes, and the recent discovery of the human non-coding RNA ANRIL, all of which expressed circular isoforms. CircRNA producing genes like the human ETS-1 gene, the human and rat cytochrome P450 genes, the rat androgen binding protein gene (Shbg), and the human dystrophin gene were also discovered.[17]
Genome-wide identification of circRNAs
Scrambled isoforms and circRNAs
In 2012, in an effort to initially identify cancer-specific exon scrambling events, scrambled exons were discovered in large numbers in both normal and cancer cells. It was found that scrambled exon isoforms comprised about 10% of the total transcript isoforms in leukocytes, with 2,748 scrambled isoforms in HeLa and H9 embryonic stem cells being identified. Additionally, about 1 in 50 expressed genes produced scrambled transcript isoforms at least 10% of the time. Tests used to recognize circularity included treating samples with RNase R, an enzyme that degrades linear but not circular RNAs, and testing for the presence of poly-A tails, which are not present in circular molecules. Overall, 98% of scrambled isoforms were found to represent circRNAs, circRNAs were found to be located in the cytoplasm, and circRNAs were found to be abundant.[17][8]
Discovery of a higher abundance of circRNAs
In 2013, a higher abundance of circRNAs was discovered. Human fibroblast RNA was treated with RNase R to enrich for circular RNAs, followed by the categorization of circular transcripts based on their abundance (low, medium, high).[3] Approximately 1 in 8 expressed genes were found to produce detectable levels of circRNAs, including those of low abundance, which was significantly higher than previously suspected, and was attributed to greater sequencing depth.[3][8]
CircRNAs tissue specificity and antagonist activity
At the same time, a computational method to detect circRNAs was developed, leading to de novo detection of circRNAs in humans, mice, and C. elegans, and extensively validating them. The expression of circRNAs was often found to be tissue/developmental stage specific. Additionally, circRNAs were found to have the ability to act as antagonists of miRNAs, microRNAs which interfere with translation of mRNAs, as exemplified by the circRNA CDR1as, which has miRNA binding sites (as seen below).[18]
CircRNAs and ENCODE Ribozero RNA-seq data
In 2014, human circRNAs were identified and quantified from ENCODE Ribozero RNA-seq data. Most circRNAs were found to be minor splice isoforms and to be expressed in only a few cell types, with 7,112 human circRNAs having circular fractions (the fraction of similarity an isoform has to transcripts the same locus) of at least 10%. CircRNAs were also found to be no more conserved than their linear controls and, according to ribosome profiling, are not translated.<[9] As previously noted, circRNAs have the ability to act as antagonists of miRNA, which is also known as the potential to act as microRNA sponges. Aside from CDR1as, very few circRNAs have the potential to act as microRNA sponges. As a whole, the majority of circular RNAs were found to be inconsequential side-products of imperfect splicing.[18][9]
CircRNAs and CIRCexplorer
In the same year, CIRCexplorer, a tool used to identify thousands of circRNAs in humans without RNase R RNA-seq data, was developed. The vast majority of identified highly expressed exonic circular RNAs were found to be processed from exons located in the middle of RefSeq genes, suggesting that the circular RNA formation is generally coupled to RNA splicing. It was determined that most circular RNAs contain multiple, most commonly, two to three, exons. Exons from circRNAs with only one circularized exon were found to be much longer than those from circRNAs with multiple circularized exons, indicating that processing may prefer a certain length to maximize exon(s) circularization. The introns of circularized exons generally contain high Alu densities that can form inverted repeated Alu pairs (IRAlus). IRAlus, either convergent or divergent, are juxtaposed across flanking introns of circRNAs in a parallel way with similar distances to adjacent exons. IRAlus, and other non-repetitive, but complementary, sequences were also found to promote circular RNA formation. On the other hand, exon circularization efficiency was determined to be affected by the competition of RNA pairing, such that alternative RNA pairing, and its competition, leads to alternative circularization. Finally, both exon circularization and its regulation were found to be evolutionarily dynamic.[15]
Genome-wide calling of circRNA in Alzheimer disease cases
Alzheimer disease (AD) cases demonstrated the role of circRNAs in health and disease. A pipeline for calling circRNA from human ribo-depleted RNA-seq both optimized and validated circRNAs' detections. An association between circRNAs and neurodegenerative diseases like AD and clinical dementia was elucidated, with a total of 148 circRNAs being significantly correlated with clinical dementia ratings at expiration/death (CDR) after false discovery rate (FDR) correction. The expression of circRNAs was independent of the lineal form and that circRNA expression was also corrected by cell proportion. CircRNAs were also found to be co-expressed with known causal Alzheimer genes, such as APP and PSEN1, indicating that some circRNAs are also part of the causal pathway. Altogether, circRNA brain expression was found to explain more about Alzheimer’s clinical manifestations than the number of APOε4 alleles, suggesting that circRNAs could be used as a potential biomarker for Alzheimer’s.[19]
Classes of CircRNA
Circular RNAs can be separated into five classes:[20][21]
| Classes of Circular RNAs | Description |
| Viroids and the hepatitis delta virus (HDV) | In viroids and HDV, single-stranded circRNAs are vital in RNA replication. Circularity allows for one initiation event to lead to multiple genomic copies in a process otherwise known as rolling circle RNA replication.[22][23][24] |
| CircRNAs from introns | Circular molecules are produced by introns produced from spliceosomal splicing, tRNA splicing, and group I and group II (self-splicing ribozymes) introns. Group I introns form circRNAs through autocatalytic ribozymal action, and while they can be detected in vivo, their function is yet to be determined.[22][23][24] Group II introns also generate circRNAs in vivo. Circular introns produced by eukaryotic spliceosomal splicing are circularized intron lariats known as circular intronic RNAs (ciRNAs). Due to circularization, ciRNAs can avoid degradation and are believed to be highly overrepresented. CiRNA function is currently unknown; however, it is speculated they may play a role in enhancing the transcription of genes they are produced from, as they interact with RNA polymerase II.[21] |
| CircRNAs from intermediates in RNA processing reactions | These are first spliced from precursors as linear molecules and then circularized with a ligase. They are essential in allowing for the rearrangement in RNA sequence order and vital in the biogenesis of permuted tRNA genes in certain algae and archaea.[21] |
| Noncoding circRNAs in archaea | Certain archaeal species have circRNAs that are produced from excised circularized tRNA introns. Circularization of functional noncoding RNAs is thought to work as a protective mechanism against exonucleases and to promote proper folding.[21][3] |
| CircRNAs in eukaryotes produced by back-splicing | Circular RNAs produced by back-splicing (a form of exon scrambling) occur when a 5′ splice site is joined to an upstream 3′ splice site. Currently, more than 25,000 different circRNAs have been identified in humans.[21][3] |
Length of circRNAs
A recent study of human circRNAs revealed that these molecules are usually composed of 1–5 exons.[18] Each of these exons can be up to three times longer than the average expressed exon,[3] suggesting that exon length may play a role in deciding which exons to circularize. 85% of circularized exons overlap with exons that code for protein,[18] although the circular RNAs themselves do not appear to be translated. During circRNA formation, exon 2 is often the upstream "acceptor" exon.[8]
Introns surrounding exons that are selected to be circularized are, on average, up to three times longer than those not flanking pre-circle exons,[8][3] although it is not yet clear why this is the case. Compared to regions not resulting in circles, these introns are much more likely to contain complementary inverted Alu repeats, Alu being the most common transposon in the genome.[3] By the Alu repeats base pairing to one another, it has been proposed that this may enable the splice sites to find each other, thus facilitating circularization.[10][3]
Introns within the circRNAs are retained at a relatively high frequency (~25%),[9] thus adding extra sequence to the mature circRNAs.
Location of circRNAs in the cell
In the cell, circRNAs are predominantly found in the cytoplasm, where the number of circular RNA transcripts derived from a gene can be up to ten times greater than the number of associated linear RNAs generated from that locus. It is unclear how circular RNAs exit the nucleus through a relatively small nuclear pore. Because the nuclear envelope breaks down during mitosis, one hypothesis is that the molecules exit the nucleus during this phase of the cell cycle.[3] However, certain circRNAs, such as CiRS-7/CDR1as, are expressed in neuronal tissues,[18][25] where mitotic division is not prevalent.
CircRNAs are stable compared to linear RNAs
CircRNAs lack a polyadenylated tail and, therefore, are predicted to be less prone to degradation by exonucleases. In 2015, Enuka et al. measured the half-lives of 60 circRNAs and their linear counterparts expressed from the same host gene and revealed that the median half-life of circRNAs of mammary cells (18.8 to 23.7 hours) is at least 2.5 times longer than the median half-life of their linear counterparts (4.0 to 7.4 hours).[26] Generally, the lifetime of RNA molecules defines their response time.[27] Accordingly, it was reported that mammary circRNAs respond slowly to stimulation by growth factors.[26]
Plausible functions of circular RNA
Evolutionary conservation of circularization mechanisms and signals
CircRNAs have been identified in various species across the domains of life. In 2011, Danan et al. sequenced RNA from Archaea. After digesting total RNA with RNase R, they were able to identify circular species, indicating that circRNAs are not specific to eukaryotes.[28] However, these archaeal circular species are probably not made via splicing, suggesting that other mechanisms to generate circular RNA likely exist.
CircRNAs were found to be largely conserved between human and sheep. By analyzing total RNA sequencing data from sheep's parietal lobe cortex and peripheral blood mononuclear cells it was shown that 63% of the detected circRNAs are homologous to known human circRNAs.[29]
In a closer evolutionary connection, a comparison of RNA from mouse testes vs. RNA from a human cell found 69 orthologous circRNAs. For example, both humans and mice encode the HIPK2 and HIPK3 genes, two paralogous kinases which produce a large amount of circRNA from one particular exon in both species.[3] Evolutionary conservation reinforces the likelihood of a relevant and significant role for RNA circularization.
CDR1as/CiRS-7 as a miR-7 sponge
microRNAs (miRNAs) are small (~21nt) non-coding RNAs that repress translation of messenger RNAs involved in a large, diverse set of biological processes.[30] They directly base-pair to target messenger RNAs (mRNAs), and can trigger cleavage of the mRNA depending on the degree of complementarity.
MicroRNAs are grouped in "seed families." Family members share nucleotides 2–7, known as the seed region.[31] Argonaute proteins are the "effector proteins" which help miRNAs carry out their job, while microRNA sponges are RNAs that "sponge up" miRNAs of a particular family, thereby serving as competitive inhibitors that suppress the ability of the miRNA to bind its mRNA targets, thanks to the presence of multiple binding sites that recognize a specific seed region.[31] Certain circular RNAs have many miRNA binding sites, which yielded a clue that they may function in sponging. Two recent papers confirmed this hypothesis by investigating a circular sponge called CDR1as/CiRS-7 in Detail, while other groups found no direct evidence for circular RNAs acting as miRNA sponges by analyzing the potential interaction of circular RNAs with the Argonaut (AGO) Protein using high-throughput sequencing of RNA isolated by cross-linking and immunoprecipitation (HITS-CLIP) data .[32]
CDR1as/CiRS-7 is encoded in the genome antisense to the human CDR1 (gene) locus (hence the name CDR1as),[18] and targets miR-7 (hence the name CiRS-7 – Circular RNA Sponge for miR-7).[25] It has over 60 miR-7 binding sites, far more than any known linear miRNA sponge.[18][25]
AGO2 is miR-7's associated Argonaute protein (see above). Though CDR1as/CiRS-7 can be cleaved by miR-671 and its associated Argonaute protein,[25] it cannot be cleaved by miR-7 and AGO2. MicroRNA cleavage activity depends on complementarity beyond the 12th nucleotide position; none of CiRS-7's binding sites meet this requirement.
An experiment with zebrafish, which do not have the CDR1 locus in their genome, provides evidence for CiRS-7's sponge activity. During development, miR-7 is strongly expressed in the zebrafish brain. To silence miR-7 expression in zebrafish, Memczak and colleagues took advantage of a tool called morpholino, which can base pair and sequester target molecules.[33] Morpholino treatment had the same severe effect on midbrain development as ectopically expressing CiRS-7 in zebrafish brains using injected plasmids. This indicates a significant interaction between CiRS-7 and miR-7 in vivo.[18]
Another notable circular miRNA sponge is SRY. SRY, which is highly expressed in murine testes, functions as a miR-138 sponge.[25][34] In the genome, SRY is flanked by long inverted repeats (IRs) over 15.5 kilobases (kb) in length. When one or both of the IRs are deleted, circularization does not occur. It was this finding that introduced the idea of inverted repeats enabling circularization.[35]
Because circular RNA sponges are characterized by high expression levels, stability, and a large number of miRNA binding sites, they are likely to be more effective sponges than those that are linear.[10]
Other possible functions for circRNAs
Though recent attention has been focused on circRNA's "sponge" functions, scientists are considering several other functional possibilities as well. For example, some areas of the mouse adult hippocampus show expression of CiRS-7 but not miR-7, suggesting that CiRS-7 may have roles that are independent of interacting with the miRNA.[18]
Potential roles include the following:
- Binding to RNA-binding proteins (RBPs) and RNAs besides miRNAs to form RNA-protein complexes.[10] These complexes could regulate RBP & RNA interactions with, for example, the canonical linear transcript of the gene.[8]
- Protein production
- Chen and Sarnow 1995 showed that a synthetic circRNA that contained an IRES (internal ribosome entry site) produced a protein product in vitro, whereas that without an IRES did not. Although the tested circRNA was a purely artificial construct, Chen and Sarnow stated in their paper that they would be interested to see whether circles naturally contain IRES elements.[36]
- Jeck et al. 2013: Tested natural circRNAs that contained a translation "start codon." However, none of these molecules bound to ribosomes, suggesting that many circRNAs may not be translated in vivo.[3]
- Transporting miRNAs inside the cell. The fact that CiRS-7 can be sliced by miR-671 might indicate the existence of a system to release a "load" of miRNAs at the appropriate time.[37]
- Regulating mRNA in the cell through limited base pairing. It is formally possible that miR-7 moderates CiRS-7's regulatory activity instead of the other way around![18][37]
Circular intronic long non-coding RNAs (ciRNAs)
Usually, intronic lariats (see above) are debranched and rapidly degraded. However, a debranching failure can lead to the formation of circular intronic long non-coding RNAs, also known as ciRNAs.[38] CiRNA formation, rather than being a random process, seems to depend on the presence of specific elements near the 5' splice site and the branchpoint site (see above).
CiRNAs are distinct from circRNAs in that they are prominently found in the nucleus rather than the cytoplasm. In addition, these molecules contain few (if any) miRNA binding sites. Instead of acting as sponges, ciRNAs seem to function in regulating the expression of their parent genes. For example, a relatively abundant ciRNA called ci-ankrd52 positively regulates Pol II transcription. Many ciRNAs remain at their "sites of synthesis" in the nucleus. However, ciRNA may have roles other than simply regulating their parent genes, as ciRNAs do localize to additional sites in the nucleus other than their "sites of synthesis".[38]
Circular RNA and disease
As with most topics in molecular biology, it is important to consider how circular RNA can be used as a tool to help mankind. Given its abundance, evolutionary conservation, and potential regulatory role, it is worthwhile to look into how circular RNA can be used to study pathogenesis and devise therapeutic interventions. For example:
- Circular ANRIL (cANRIL) is the circular form of ANRIL, a long non-coding RNA (ncRNA). Expression of cANRIL is correlated with risk for atherosclerosis, a disease in which the arteries become hard. It has been proposed that cANRIL can modify INK4/ARF expression, which, in turn, increases risk for atherosclerosis.[39] Further study of cANRIL expression could potentially be used to prevent or treat atherosclerosis.
- miR-7 plays an important regulatory role in several cancers and in Parkinson's disease, which is a brain illness of unknown origin.[25] Perhaps CiRS-7's sponge activity could help in countering miR-7 activity. If circular sponge activity can indeed help in countering harmful miRNA activity, scientists will need to figure out the best way to introduce sponge expression, perhaps via a transgene, which is a synthetic gene that is transferred between organisms. It is also important to consider how transgenes can be expressed only in specific tissues, or expressed only when induced.[31]
- Circular RNAs were found to be regulated by hypoxia, especially the circRNA cZNF292 was found to have proangiogenic activities in endothelial cells.[32]
Circular RNAs play a role in Alzheimer disease pathogenesis
Dube et al.,[19] demonstrated for the first time that brain circular RNAs (circRNA) are part of the pathogenic events that lead to Alzheimer’s disease, hypothesizing that specific circRNA would be differentially expressed in AD cases compared to controls and that those effects could be detected early in the disease. They optimized and validated a novel analyses pipeline for circular RNAs (circRNA). They performed a three-stage study design, using the Knight ADRC brain RNA-seq data as discovery (stage 1), using the data from Mount Sinai as replication (stage 2) and a meta-analysis (stage 3) to identify the most significant circRNA differentially expressed in Alzheimer disease. Using his pipeline, they found 3,547 circRNA that passed stringent QC in the Knight ADRC cohort that includes RNA-seq from 13 controls and 83 Alzheimer cases, and 3,924 circRNA passed stringent QC in the MSBB dataset. A meta-analysis of the discovery and replication results revealed a total of 148 circRNAs that were significantly correlated with CDR after FDR correction. In addition, 33 circRNA passed the stringent gene-based, Bonferroni multiple test correction of 5×10-6, including circHOMER1 (P =2.21×10−18) and circCDR1-AS (P = 2.83 × 10−8), among others. They also performed additional analyses to demonstrate that the expression of circRNA were independent of the lineal form as well as the cell proportion that can confound the brain RNA-seq analyses in Alzheimer disease studies. They performed co-expression analyses of all the circRNA together with the lineal forms and found that circRNA, including those that were differentially expressed in Alzheimer disease compared to controls co-expressed with known causal Alzheimer genes, such as APP and PSEN1, indicating that some circRNA are also part of the causal pathway. They also demonstrated that cirRNA brain expression explained more about Alzheimer clinical manifestations that the number of APOε4 alleles, suggesting that could be used as a potential biomarker for Alzheimer disease. This is an important study for the field, as it is the first time that circRNA are quantified and validated (by real-time PCR) in human brain samples at genome-wide scale and in large and well-characterized cohorts. It also demonstrates that these RNA forms are likely to be implicated on complex traits including Alzheimer disease will help to understand the biological events that leads to disease.
Viroids as circular RNAs
Viroids are mostly plant pathogens, which consist of short stretches (a few hundred nucleobases) of highly complementary, circular, single-stranded, and non-coding RNAs without a protein coat. Compared with other infectious plant pathogens, viroids are extremely small in size, ranging from 246 to 467 nucleobases; they thus consist of fewer than 10,000 atoms. In comparison, the genome of the smallest known viruses capable of causing an infection by themselves are around 2,000 nucleobases long.[40]
See also
References
- ^ Dawoud, Alyaa (12 October 2022). "Circular RNAs: New layer of complexity evading breast cancer heterogeneity". Non-coding RNA Research. Retrieved 13 October 2022.
- ^ Pamudurti, Nagarjuna Reddy; Bartok, Osnat; Jens, Marvin; et al. (April 2017). "Translation of CircRNAs". Molecular Cell. 66 (1): 9–21.e7. doi:10.1016/j.molcel.2017.02.021. PMC 5387669. PMID 28344080.

- ^ a b c d e f g h i j k l m n Jeck, WR; Sorrentino, JA; Wang, K; et al. (February 2013). "Circular RNAs are abundant, conserved, and associated with ALU repeats". RNA. 19 (2): 141–157. doi:10.1261/rna.035667.112. PMC 3543092. PMID 23249747.
- ^ Vromman, Marieke; Vandesompele, Jo; Volders, Pieter-Jan (2021). "Closing the circle: current state and perspectives of circular RNA databases". Briefings in Bioinformatics. 22 (1): 288–297. doi:10.1093/bib/bbz175. PMC 7820840. PMID 31998941.
- ^ a b c d Reece, JB (2010). Campbell Biology (9th ed.). San Francisco: Benjamin Cummings.
- ^ Yu, J; Hu, S; Wang, J; Wong, GK; Li, S; Liu, B; Deng, Y; Dai, L; Zhou, Y; Zhang, X; Cao, M; Liu, J; Sun, J; Tang, J; Chen, Y; Huang, X; Lin, W; Ye, C; Tong, W; Cong, L; Geng, J; Han, Y; Li, L; Li, W; Hu, G; Huang, X; Li, W; Li, J; Liu, J; et al. (2002). "A draft sequence of the rice genome (Orya sativa L. ssp. indica)". Science. 296 (5565): 79–92. Bibcode:2002Sci...296...79Y. doi:10.1126/science.1068037. PMID 11935017. S2CID 208529258.
- ^ Celotto, A.M.; Graveley, B.R. (2001). "Alternative splicing of the Drosophila Dscam pre-mRNA is both temporally and spatially regulated". Genetics. 159 (2): 599–608. doi:10.1093/genetics/159.2.599. PMC 1461822. PMID 11606537.
- ^ a b c d e f g Salzman, J; Gawad, C.; Wang, P.L.; Lacayo, N; Brown, PO (2012). "Circular RNAs are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types". PLoS One. 7 (2): e30733. Bibcode:2012PLoSO...730733S. doi:10.1371/journal.pone.0030733. PMC 3270023. PMID 22319583.
- ^ a b c d Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. (2014). "Expanded identification and characterization of mammalian circular RNAs". Genome Biology. 15 (7): 409. doi:10.1186/s13059-014-0409-z. PMC 4165365. PMID 25070500.
- ^ a b c d Wilusz, J.E.; Sharp, PA (2013). "A Circuitous Route to Noncoding RNA" (PDF). Science. 340 (6131): 440–41. Bibcode:2013Sci...340..440W. doi:10.1126/science.1238522. PMC 4063205. PMID 23620042.
- ^ Daniel, Chammiran; Silberberg, Gilad; Behm, Mikaela; Öhman, Marie (2014-02-03). "Alu elements shape the primate transcriptome by cis-regulation of RNA editing". Genome Biology. 15 (2): R28. doi:10.1186/gb-2014-15-2-r28. ISSN 1474-760X. PMC 4053975. PMID 24485196.
- ^ Ivanov, Andranik; Memczak, Sebastian; Wyler, Emanuel; Torti, Francesca; Porath, Hagit T.; Orejuela, Marta R.; Piechotta, Michael; Levanon, Erez Y.; Landthaler, Markus; Dieterich, Christoph; Rajewsky, Nikolaus (January 2015). "Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals". Cell Reports. 10 (2): 170–177. doi:10.1016/j.celrep.2014.12.019. ISSN 2211-1247.
- ^ Wilusz, Jeremy E (2015-05-04). "Repetitive elements regulate circular RNA biogenesis". Mobile Genetic Elements. 5 (3): 39–45. doi:10.1080/2159256X.2015.1045682. PMC 4588227. PMID 26442181.
- ^ Liang, Dongming; Wilusz, Jeremy E. (2014-10-15). "Short intronic repeat sequences facilitate circular RNA production". Genes & Development. 28 (20): 2233–2247. doi:10.1101/gad.251926.114. ISSN 0890-9369. PMC 4201285. PMID 25281217.
- ^ a b Zhang, Xiao-Ou; Wang, Hai-Bin; Zhang, Yang; Lu, Xuhua; Chen, Ling-Ling; Yang, Li (September 2014). "Complementary Sequence-Mediated Exon Circularization". Cell. 159 (1): 134–147. doi:10.1016/j.cell.2014.09.001. ISSN 0092-8674.
- ^ a b c Kokot, Karoline E.; Kneuer, Jasmin M.; John, David; Rebs, Sabine; Möbius-Winkler, Maximilian N.; Erbe, Stephan; Müller, Marion; Andritschke, Michael; Gaul, Susanne; Sheikh, Bilal N.; Haas, Jan (2022-06-23). "Reduction of A-to-I RNA editing in the failing human heart regulates formation of circular RNAs". Basic Research in Cardiology. 117 (1): 32. doi:10.1007/s00395-022-00940-9. ISSN 1435-1803.
- ^ a b Barrett, Steven P.; Salzman, Julia (2016-06-01). "Circular RNAs: analysis, expression and potential functions". Development. 143 (11): 1838–1847. doi:10.1242/dev.128074. ISSN 0950-1991. PMC 4920157. PMID 27246710.
- ^ a b c d e f g h i j Memczak, Sebastian; Jens, Marvin; Elefsinioti, Antigoni; Torti, Francesca; Krueger, Janna; Rybak, Agnieszka; Maier, Luisa; Mackowiak, Sebastian D.; Gregersen, Lea H.; Munschauer, Mathias; Loewer, Alexander (March 2013). "Circular RNAs are a large class of animal RNAs with regulatory potency". Nature. 495 (7441): 333–338. Bibcode:2013Natur.495..333M. doi:10.1038/nature11928. ISSN 1476-4687. PMID 23446348. S2CID 4416605.
- ^ a b Dube, Umber; Del-Aguila, Jorge L.; Li, Zeran; Budde, John P.; Jiang, Shan; Hsu, Simon; Ibanez, Laura; Fernandez, Maria Victoria; Farias, Fabiana; Norton, Joanne; Gentsch, Jen (November 2019). "An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations". Nature Neuroscience. 22 (11): 1903–1912. doi:10.1038/s41593-019-0501-5. ISSN 1546-1726. PMC 6858549. PMID 31591557.
- ^ Eger, Nicole; Schoppe, Laura; Schuster, Susanne; Laufs, Ulrich; Boeckel, Jes-Niels (2018), Xiao, Junjie (ed.), "Circular RNA Splicing", Circular RNAs, Singapore: Springer Singapore, vol. 1087, pp. 41–52, doi:10.1007/978-981-13-1426-1_4, ISBN 978-981-13-1425-4, retrieved 2022-06-24
- ^ a b c d e Nisar, Sabah; Bhat, Ajaz A.; Singh, Mayank; Karedath, Thasni; Rizwan, Arshi; Hashem, Sheema; Bagga, Puneet; Reddy, Ravinder; Jamal, Farrukh; Uddin, Shahab; Chand, Gyan (2021-02-05). "Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies". Frontiers in Cell and Developmental Biology. 9: 617281. doi:10.3389/fcell.2021.617281. ISSN 2296-634X. PMC 7894079. PMID 33614648.
- ^ a b Grabowski, P. J.; Zaug, A. J.; Cech, T. R. (February 1981). "The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena". Cell. 23 (2): 467–476. doi:10.1016/0092-8674(81)90142-2. ISSN 0092-8674. PMID 6162571. S2CID 41905828.
- ^ a b Kruger, K.; Grabowski, P. J.; Zaug, A. J.; Sands, J.; Gottschling, D. E.; Cech, T. R. (November 1982). "Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena". Cell. 31 (1): 147–157. doi:10.1016/0092-8674(82)90414-7. ISSN 0092-8674. PMID 6297745. S2CID 14787080.
- ^ a b Zaug, A. J.; Grabowski, P. J.; Cech, T. R. (17–23 February 1983). "Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction". Nature. 301 (5901): 578–583. Bibcode:1983Natur.301..578Z. doi:10.1038/301578a0. ISSN 0028-0836. PMID 6186917. S2CID 4348876.
- ^ a b c d e f Hansen, T.B.; Jensen, TI; Clausen, BH; Bramsen, JB; Finsen, B; Damgaard, CK; Kjems, J (2013). "Natural RNA circles function as efficient microRNA sponges". Nature. 495 (7441): 384–88. Bibcode:2013Natur.495..384H. doi:10.1038/nature11993. PMID 23446346. S2CID 205233122.
- ^ a b Enuka, Y.; Lauriola, M; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. (2015). "Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor". Nucleic Acids Research. 44 (3): 1370–83. doi:10.1093/nar/gkv1367. PMC 4756822. PMID 26657629.
- ^ Sneppen, K (2014). Models of Life - Dynamics and Regulation in Biological Systems. San Francisco: Cambridge University Press. ISBN 978-1-107-06190-3.
- ^ Danan, M; Schwartz, S; Edelheit, S; Sorek, R (2012). "Transcriptome-wide discovery of circular RNAs in Archaea". Nucleic Acids Research. 40 (7): 3131–42. doi:10.1093/nar/gkr1009. PMC 3326292. PMID 22140119.
- ^ Varela-Martínez, E; Corsi, GI; Anthon, C; Gorodkin, J; Jugo, BM (2021). "Novel circRNA discovery in sheep shows evidence of high backsplice junction conservation". Scientific Reports. 11 (427): 427. doi:10.1038/s41598-020-79781-2. PMC 7801505. PMID 33432020.
- ^ Ding, XC; Weiler, J; Grosshans, H (2009). "Regulating the regulators: mechanisms controlling the maturation of microRNAs". Trends in Biotechnology. 27 (1): 27–36. doi:10.1016/j.tibtech.2008.09.006. PMID 19012978.
- ^ a b c Ebert, MS; Sharp, PA (2010). "MicroRNA sponges: progress and possibilities". RNA. 16 (11): 2043–50. doi:10.1261/rna.2414110. PMC 2957044. PMID 20855538.
- ^ a b Boeckel, Jes-Niels; Jaé, Nicolas; Heumüller, Andreas W.; Chen, Wei; Boon, Reinier A.; Stellos, Konstantinos; Zeiher, Andreas M.; John, David; Uchida, Shizuka (2015-10-23). "Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA". Circulation Research. 117 (10): 884–890. doi:10.1161/CIRCRESAHA.115.306319. ISSN 1524-4571. PMID 26377962.
- ^ Summerton, J (1999). "Morpholino antisense oligomers: the case for an RNase H-independent structural type". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1489 (1): 141–58. doi:10.1016/S0167-4781(99)00150-5. PMID 10807004.
- ^ Capel, B; Swain, A; Nicolis, S; Hacker, A; Walter, M; Koopman, P; Goodfellow, P; Lovell-Badge, R (1993). "Circular transcripts of the testis-determining gene Sry in adult mouse testis". Cell. 73 (5): 1019–30. doi:10.1016/0092-8674(93)90279-y. PMID 7684656.
- ^ Dubin, RA; Kazmi, MA; Ostrer, H (1995). "Inverted repeats are necessary for circularization of the mouse testis Sry transcript". Gene. 167 (1–2): 245–48. doi:10.1016/0378-1119(95)00639-7. PMID 8566785.
- ^ Chen, CY; Sarnow, P (1995). "Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs". Science. 268 (5209): 415–17. Bibcode:1995Sci...268..415C. doi:10.1126/science.7536344. PMID 7536344.
- ^ a b Hentze, MW; Preiss, T (2013). "Circular RNAs: splicing's enigma variations". The EMBO Journal. 32 (7): 923–25. doi:10.1038/emboj.2013.53. PMC 3616293. PMID 23463100.
- ^ a b Zhang, Y; Zhang, XO; Chen, T; Xiang, JF; Yin, QF; Xing, YH; Zhu, S; Yang, L; Chen, LL (2013). "Circular Intronic Long Non-coding RNAs". Molecular Cell. 51 (6): 1–15. doi:10.1016/j.molcel.2013.08.017. PMID 24035497.
- ^ Burd, CE; Jeck, WR; Liu, Y; Sanoff, HK; Wang, Z; Sharpless, NE (2010). "Expression of Linear and Novel Circular Forms of an INK4/ARF-Associated Non-coding RNA Correlates with Atherosclerosis Risk". PLOS Genetics. 6 (12): e1001223. doi:10.1371/journal.pgen.1001233. PMC 2996334. PMID 21151960.
- ^ Sanger, H. L.; Klotz, G.; Riesner, D.; Gross, H. J.; Kleinschmidt, A. K. (1 November 1976). "Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures". Proceedings of the National Academy of Sciences. 73 (11): 3852–3856. Bibcode:1976PNAS...73.3852S. doi:10.1073/pnas.73.11.3852. PMC 431239. PMID 1069269.